The coronavirusCOVID-19 is affecting 210 countries and territories. Till now the death toll rises to 1,68,784 (date: 20.04.2020). However, the world has already seen a similar pandemic in the year 2002 by a similar type of coronavirus SARS-CoV. The similarities between SARS-CoV and SARS-CoV-2 are striking, not only in name but also in their action. The whole genome of SARS-CoV-2 has an 86% similarity with SARS-CoV. Both viruses share high degrees of homology to SARS-like coronaviruses isolated in bats, suggesting that bats are the probable origin of both SARS-CoV and SARS-CoV-2. However, there are certain key differences have been observed that make the SARS-CoV-2 is a global health crisis.
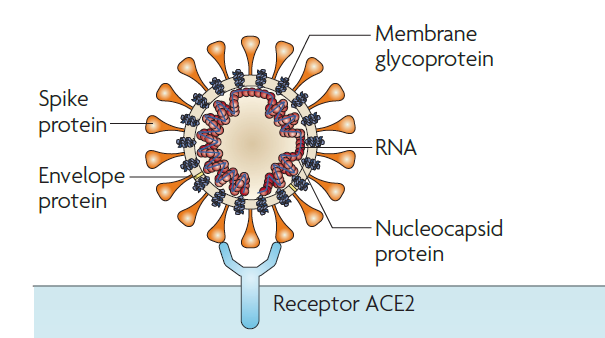
Here we will first describe how this virus attaches to the host cell and enters the host cell. Coronavirus entry into the host cell is mediated by the transmembrane spike (S) protein residing on the virion surface. One subunit of S protein recognizes the host cell receptor. All SARS related coronaviruses interact directly with angiotensin-converting enzyme 2 (ACE2). Figure 1 shows how the membrane-bound S protein binds to the host cell receptor. The interaction between the S protein and the receptor ACE2 facilitates the entry of the virus into the host cell. As the S protein binding to the ACE2 is the first step toward the host cell recognition, this step is also the target for the therapeutics and vaccine design. In humans, S protein binds to the human angiotensin-converting enzyme 2 (hACE2). The published data shows that SARS-CoV-2 binds almost five times more effectively with the hACE2 as compared to the SARS-CoV.
Bioinformatic analysis of the S protein of SARS-CoV-2 and SARS-CoV identifies the key residues that are responsible for efficient binding to the hACE2 protein. The recent crystal structure of SARS-CoV-2 also elucidates key differences between the binding domain of 2019 SARS and 2002 SARS coronaviruses.
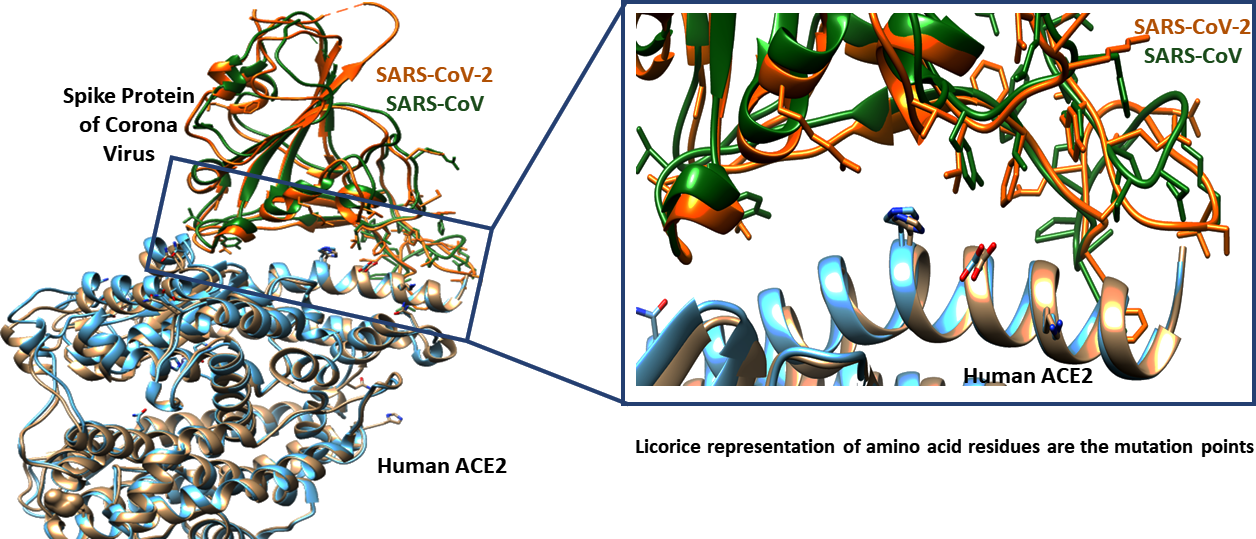
Superposition of receptor-binding domain of S protein of SARS-CoV-2 (PDB ID: 6VW1) and SARS-CoV shows very similar structural fold (Figure 2). However, there are 26 mutation points observed in SARS-CoV-2 receptor binding domain.
Figure 2: Superimposition of SARS-CoV-2 and SARS-CoV with ACE2. The interface region is highlighted inside the box. The crystal structures that are used for structural alignment of SARS-CoV-2 and SAR-CoV are 6VW1 and 2AJF respectively
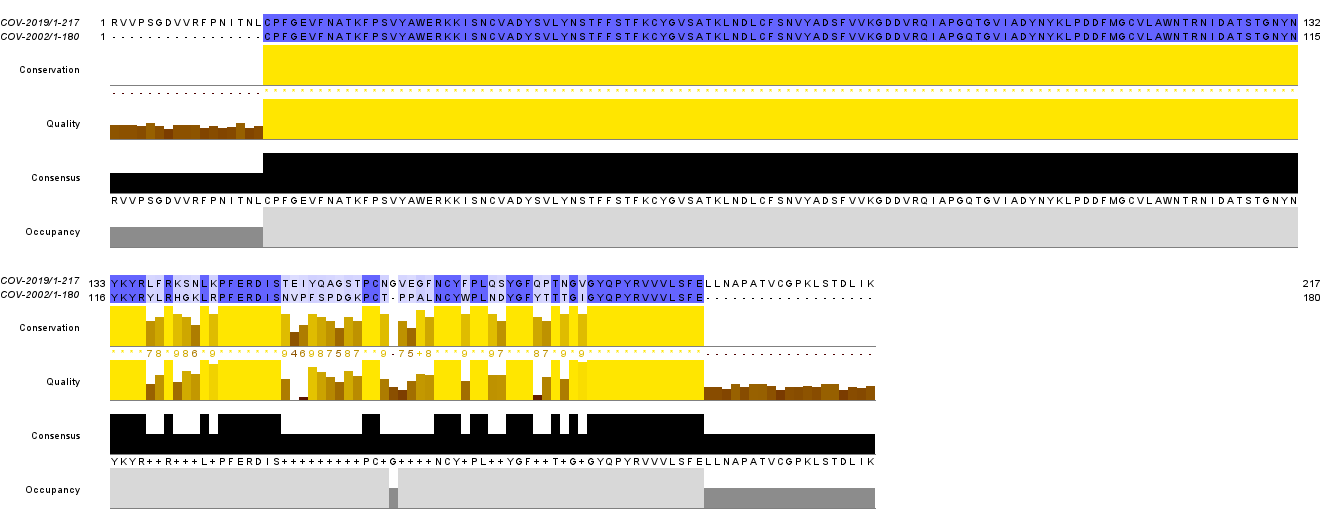
Sequence alignment shows the mutation points between the SARS-CoV-2 and SARS-CoV (Figure 3). The important alternations are Val404 to Lys417, Arg426to Asn439, Tyr442 to Leu455, Leu443 to Phe456, Tyr484 to Gln498, Thr487 to Asn501, etc. The substitution at the interface leads to the change in affinity towards ACE2. The variation near the interface domain of S and ACER protein suggests enhancement in binding affinity for SARS-CoV2. This may be one possible reason why SARS-2019 is deadlier than SARS-2002.
Figure 3: Sequence alignment of SARS-CoV-2 and SARS-CoV. The blue line represents the conserved residues while the white space represents the mutation points. The image is created using Jalview.
References
- Yan, R. Zhang, Y. Li, Y. Xia, L. Guo, Y. Zhou, Q. Structural Basis for the Recognition of SARS-CoV-2 by Full-Length Human ACE2. Science (80-. ). 2020 , 367 (6485), 1444 LP – 1448. https://doi.org/10.1126/science.abb2762.
- Du, L. He, Y. Zhou, Y. Liu, S. Zheng, B.-J. Jiang, S. The Spike Protein of SARS-CoV — a Target for Vaccine and Therapeutic Development. Nat. Rev. Microbiol. 2009, 7 (3), 226–236. https://doi.org/10.1038/nrmicro2090.
- Li, F. Li, W. Farzan, M. Harrison, S. C. Structure of SARS Coronavirus Spike Receptor-Binding Domain Complexed with Receptor. Science (80-. ). 2005, 309 (5742), 1864 LP – 1868. https://doi.org/10.1126/science.1116480.
- https://www.worldometers.info/coronavirus/