Understanding the sequence-function relationship of the enzymes is necessary for the rational engineering of enzymes towards their superior self. Over the years, scientists are seeking the answer: How enzyme sequence is related to its function? While experimental methods like directed evolution yield enzymes with desired enhancements, they are always hampered by the design of selective pressure especially if the enzyme is involved in the catalysis. Protein engineering and saturation mutagenesis experiments have very low chances of yielding high-performing enzymes as mutations, in general, are deleterious.
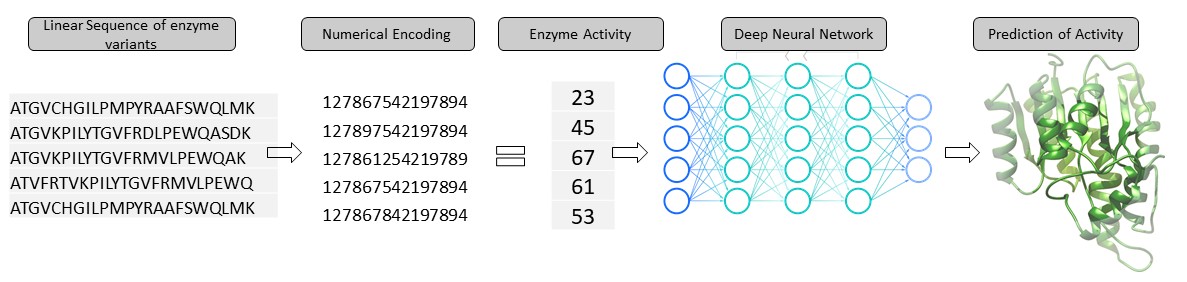
Figure 1 - Linear Sequence of amino acids dictates the three dimensional structure of protein
However, both the successful and failed attempts guide the further iterations of engineering, if the underlying sequence-function relationship is understood based on the performance of the mutants. In this respect, artificial intelligence/machine learning (AI/ML) models play a crucial role in understanding the sequence-function relationship by interpolating/extrapolating over the enzyme fitness landscape1,2
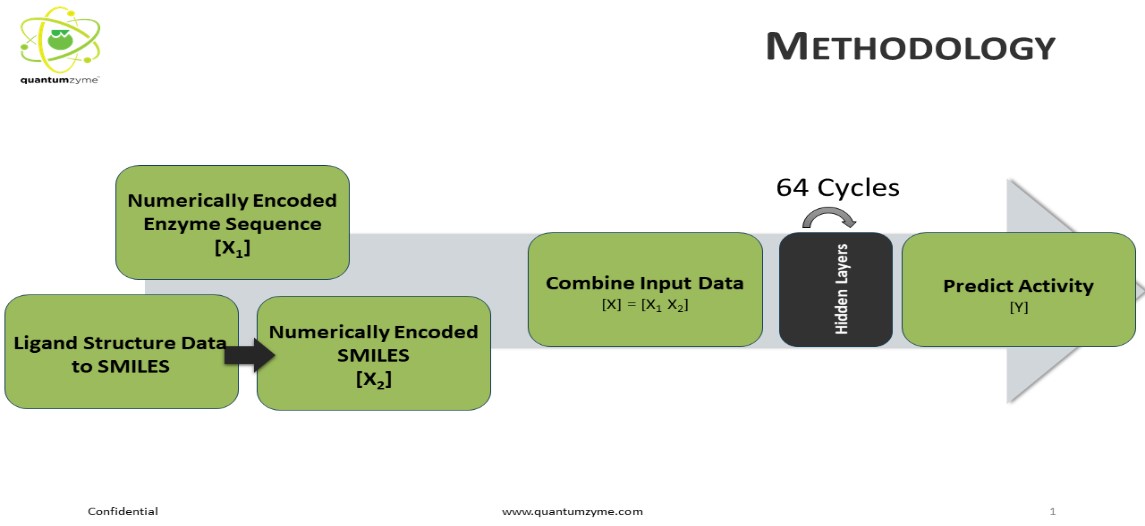
To address the problem of sequence to function, Quantumzyme has been working on developing machine learning and AI algorithms. The QZ-E001 AI/ML tool derives the input features in the simplest and most easily accessible formats without relying on complex experimental data to explore the sequence-function landscape. The prediction platform requires enzyme and ligand(s) information in the form of sequence and SMILES, respectively, along with the associated activity data. The automated pre-processing segment numerically encodes the input data feeding it into the core architecture of our tool consisting of a series of dense networks. The specialized deep-learning architecture allows the users to train and predict on various forms of activity data including, but not limited to, binding specificity (Km or KI), catalytic rate constant (KCat), enantiomer selectivity (ee% or ΔΔG), and in terms of product formation/substrate consumption (measured as % product formed or rate of product formation and vice versa). Depending on the nature of the available training dataset, our tool can be used to predict the activity of mutants even towards a novel substrate. In the upcoming releases, the model is being upgraded to assess and predict best performing mutants with a high confidence score. To train, test, and validate our AI/ML platform, we extracted the binding affinity data belonging to the protein variants against a batch of ligands with diversified physicochemical properties and the catalytic rate constant data of protein variants tested against substrates of varying size and shape available in the public domain databases. We intentionally relied on sparse datasets to train and test the model to mimic the possible industrial-case scenario. The data points are randomly divided into 70:30% of training and validation sets initially to tune the model. The underlying AI/ML model is trained using the training dataset and is blinded from the validation set.
Figure 2 - The overall flow, input and output of AI-ML model
The pre-preparatory and the prediction pipeline shown in Figure 1 are fully automated for ease of purpose. To address the issue of overfitting and diversifying the training data, Quantumzyme has worked on creating synthetic data with respect to the protein sequences, and the respective activity values. The synthetic data points add more depth to the variations in the protein sequence. Our automated pipeline recognizes and deals with the minority class in our training datasets by appropriately oversampling the data points. Our inherent synthetic data augments the original data by creating new data points without affecting the variance of the input data. To ensure that the results are not virtual and misguiding due to the added synthetic data, the model was trained on combined synthetic and original data but tested on purely the original data.
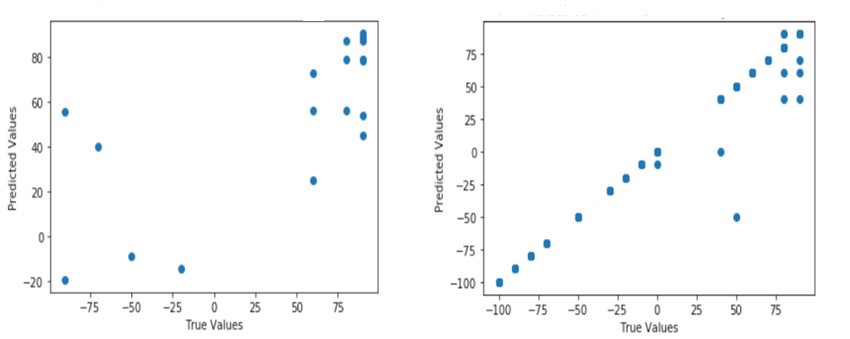
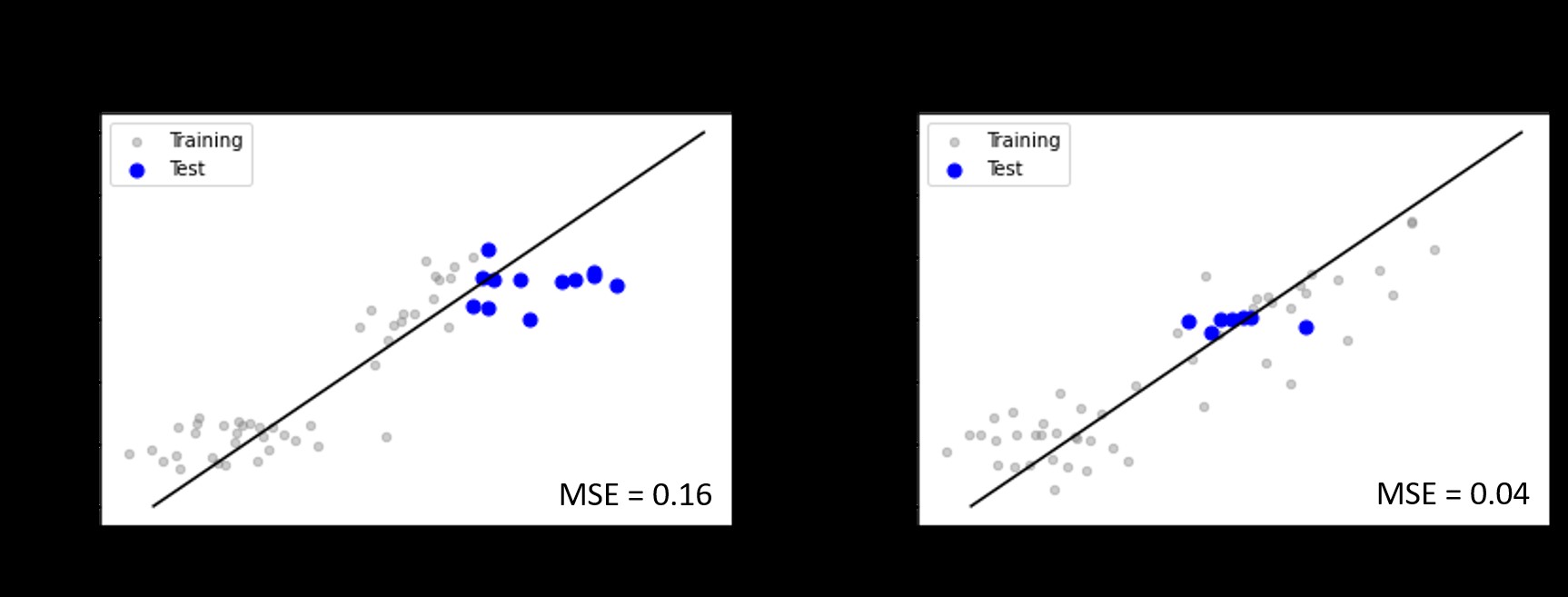
Figure 3 demonstrates the results of two different trials. First, the model was trained on original data and the predicted activity values were plotted against the true activity values. In the second trial, the model was trained on both original and synthetic data and the results were plotted. The deviation in the scatter plot for original data clearly shows the importance of having synthetic data points in the training process.
Figure 3 - Comparison of original (a) and the synthetic data points (b).
Once the core architecture is defined, we tested the performance of the model on two commercially required extreme scenarios (A & B) using the validation set (30% of the initially isolated data points). In scenario A, we simulated a case of datapoints lacking potentially high-performing enzyme variants, where all the mutants explored until then showed relatively low (or neutral) performance to that of the wild type. While in the case of scenario B, we explored an interesting case of predicting the activity of a panel of enzymes against a novel substrate using the previously known performance of the enzyme panel with conventional substrates.
Figure 4 demonstrates the performance of our tool in dealing with these commercially attractive scenarios. All the test data points fall along the 1:1 correlation line (black solid line in Figure 3) with deviations less than one standard deviation. The prediction errors, measured in terms of mean squared error (MSE), are equivalent to that of the experimental error range observed in some cases of the training set. Painstakingly, the model has been repeatedly tested on various publicly available datasets of various scenarios (one enzyme: multiple ligands, multiple enzymes: one ligand, multiple enzymes: multiple substrates, etc.,) and on diverse forms of activity measures (Km, KCat, KD, ∆∆G, and % product formed).
Finally, we will highlight some inherent challenges in an AI/ML model. Over the years, deep learning techniques create a lot of promise and success, however, the outcome that arises from algorithms is only as good as the data that go in. Albeit the tedious checkpoints placed in our automated pipeline to avoid overfitting the data, the poorly sample low confidence data points may bias the outcomes and performance of the tool. With sufficient high-quality experimental data points available in most enzyme engineering industries, the performance of the model is unchallenged with our previous efforts using conventional methods.
To summarize this blog, Quantumzyme has embarked on this journey to develop a machine learning framework to predict the properties of an enzyme with significant accuracy and we have, once again, surpassed our previous performances with this AI/ML-based enzyme engineering platform.
References
- Wittmann, B. J.; Johnston, K. E.; Wu, Z.; Arnold, F. H. Advances in Machine Learning for Directed Evolution. Curr. Opin. Struct. Biol. 2021, 69, 11–18.
Mazurenko, S.; Prokop, Z.; Damborsky, J. Machine