Introduction to enhanced sampling:
Molecular Dynamics (MD) simulation is an important technique to analyse the system at the atomic scale. The main purpose of MD simulation is to sample all the states in the system that we are interested in. Most realistic systems are characterized by the free energy surface (FES) having multiple minima separated by large free energy barriers. Sampling these states with conventional MD simulations will take days due to the limited time scale. And most biological processes like enzyme-substrate reactions require orders of magnitude longer than what is achieved in molecular dynamics simulation on a nanosecond or microsecond scale. That’s why we need improved MD techniques which can sample all the the conformational spaces. One of the most effective and widely used technique to resolve this is Enhanced MD Simulations. Enhanced MD simulations define the system by set of collective variables. External potential is used to bias the system so that it can overcome the local minima by crossing the free energy barrier. Thus, it can sample the whole conformational space. Two widely used techniques for enhanced sampling MD are Umbrella sampling and Metadynamics.
- Umbrella Sampling1-2: It is widely used enhanced sampling technique which uses harmonic potential as the external potential for sampling. It discretises the whole sample space into series of small windows and then applies a potential which acts as a restraint force so that the reaction coordinate remains close to the centre of the window. And then classical MD simulation is carried out in each window independently. Free energy is calculated across each window and then respective free energy plots from each window are stitched together with the help of mathematical equations model weighted histogram analysis (WHAM) to get the final free energy plot of the whole sample space.
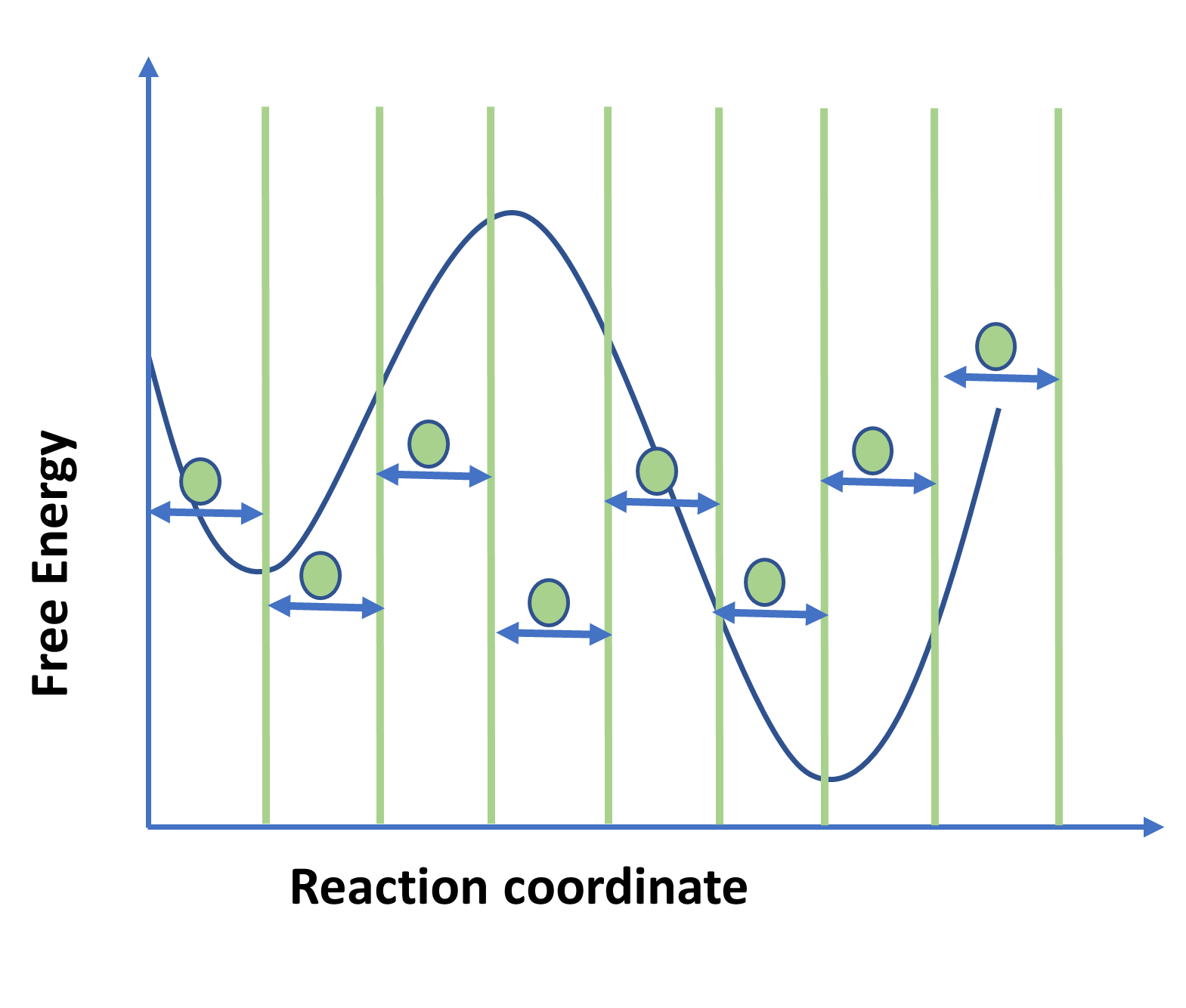
Figure 1: Schematic diagram showing free energy plot using Umbrella sampling.
Figure represent discretization of reaction coordinate space in multiple windows separated by green vertical lines. Enhanced sampling methods are used to accelerate molecular dynamics (MD) to quickly explore the conformational space. Most important factor involved[RS1] [IB2] in enhanced sampling is the reaction coordinate which describes the system and free energy that is plotted as a function of the reaction coordinate. Choosing the reaction coordinate becomes crucial in enhanced techniques.
- Metadynamics3-4: This enhanced sampling method uses external history dependent potential to the system which helps to sample the conformational spaces which were not explored by classical MD simulation. External potential restricts system to go back to previously visited states which helps system to sample high energy regions also.
A
B
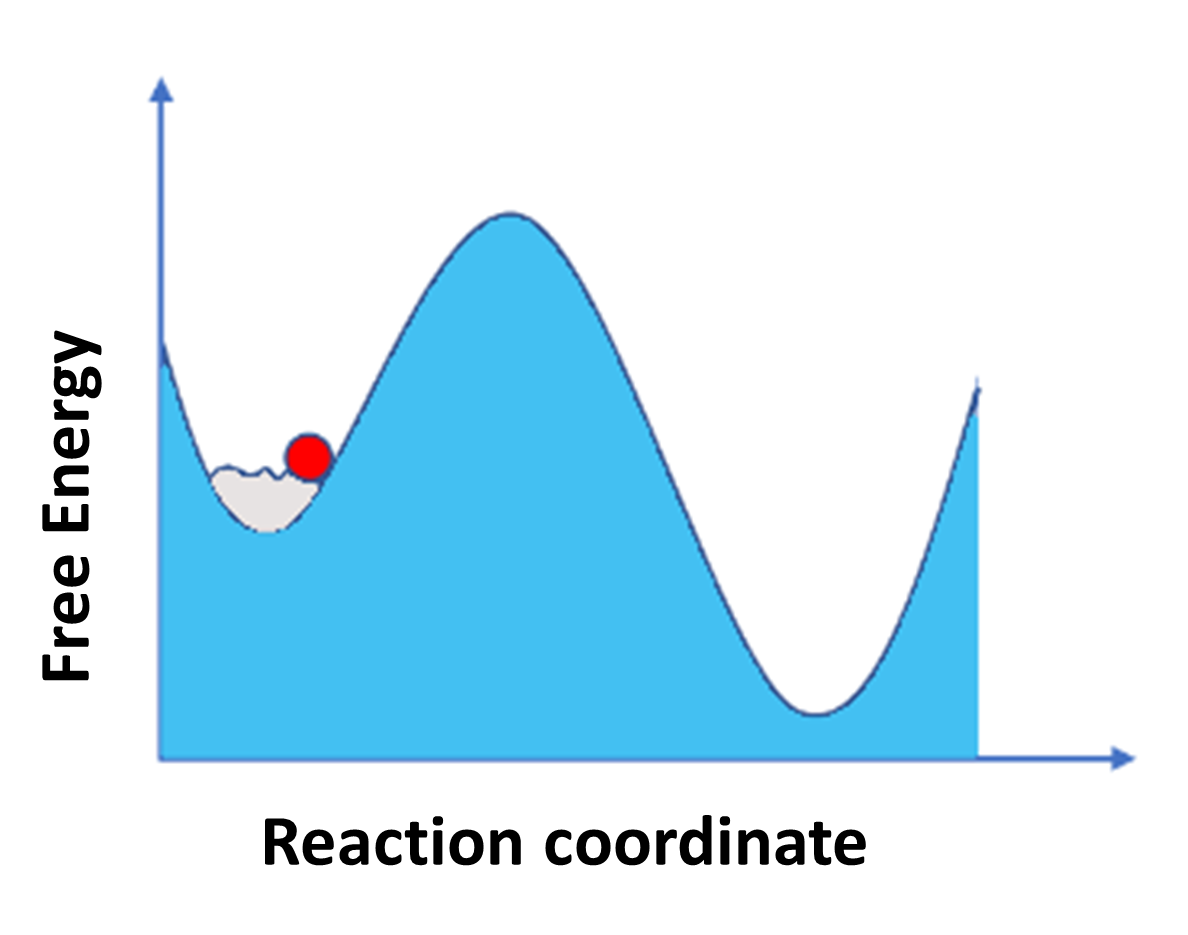
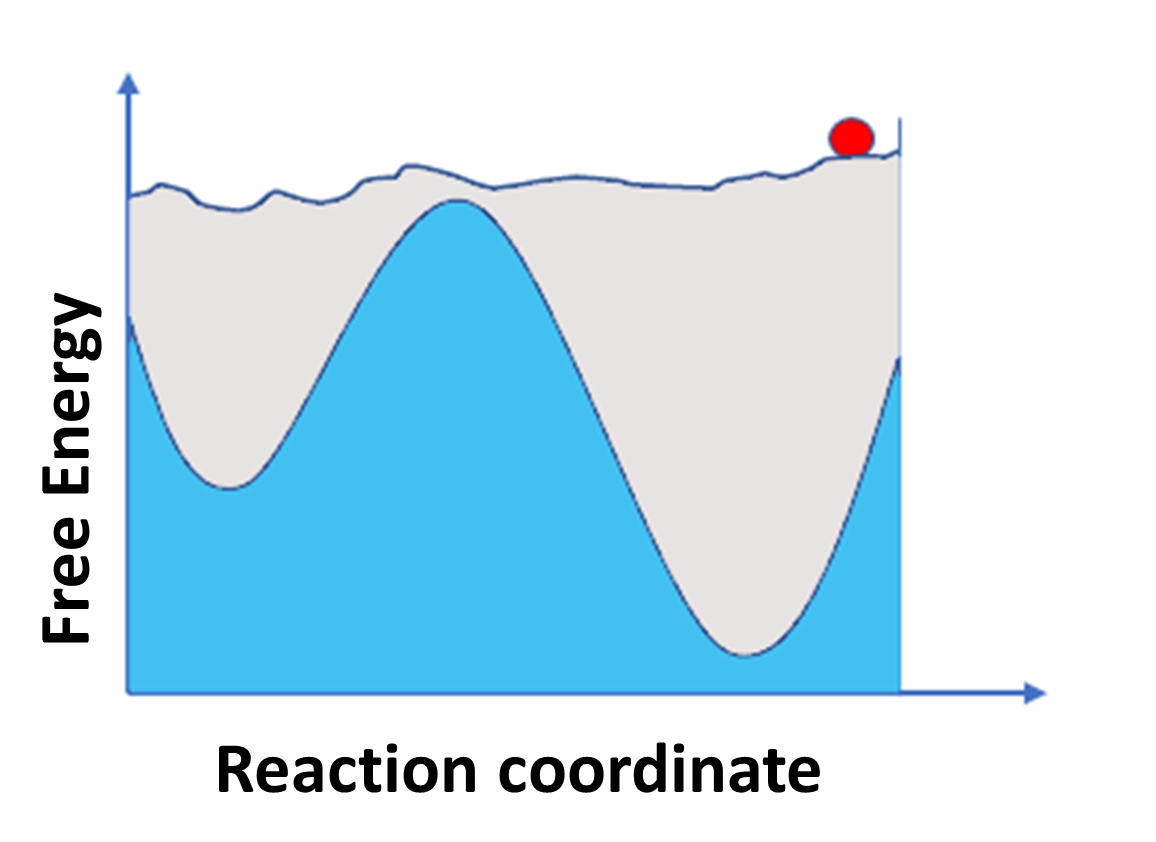
Figure 2: (A) representation of classical MD simulation and (B) represent Enhanced sampling MD simulation.
Figure shows classical MD simulation on the left side where particle is stuck in minima and it is not able to explore other region due to high energy barrier while on the right-side (B) enhanced MD simulation technique is used where particle is pushed by external potential so that it is able to sample all spaces.
Selection of reaction coordinate:
Reaction coordinate is a continuous function which is sufficient to define your system. The quality and reliability of the results obtained from enhanced sampling techniques are mostly dependent on selections which is also referred to as collective Variable (CV). The assumption taken in all the above-mentioned enhanced sampling methods is that the chosen CV must cover all the dynamics in the system, and if some motion is not captured by the CV, must be of fast dynamics. The success of the enhanced sampling is on how to choose the proper low dimension CV or CVs. We can select many simple CVs to sample the whole conformational space such as distance, angle, Coordination number, etc. Most of the tutorials on enhanced sampling have worked on the dihedral angle for the alanine di-peptide system. Selection of CV depends on your system and phenomenon you are studying, for e.g., when we are looking for binding[IB1] free energy of substrate and enzyme then distance and dihedral can be chosen as CV.
Limitation of CV:
CV/CVs can be very simple like distance, coordination number, dihedral, or an arbitrary combination. Simpler CVs like distance, coordination number, or dihedral are more useful to understand and also easy to build the free energy surface. But for many CVs, we used to get an artifact and the final free energy landscape cannot be trusted fully. For example, if we choose RMSD as the CV, it is highly degenerate. For one single value of RMSD, there can be many more conformations, and also at high RMSD, we will get overestimated minima that may not be the actual minima. Similarly, coordination numbers also can give inaccurate FES for particular systems because contacts are easy to break and difficult to form. Therefore, at zero coordination number, we will get an artificial minimum. Hence the trickiest part is choosing of CV/CVs which works for their system of interest.
Video shows unbinding of ligand from active site to solvent medium
The cofactor available in the enzyme are shown in pink colour, ligand is in green colour and Protein chains are represented in cyan.
Role of enhanced sampling in Enzyme catalysis.
For most enzyme-substrate reactions, the binding of the substrate in the active site of the enzyme is very important. From our practical experience in Quantumzyme, we have noticed that the active site is not always at the enzyme surface, in most of the projects we work on, we found binding pocket is through a tunnel in a deep pocket. To understand the enzyme-substrate binding or to explore the activity of the enzyme towards the substrate, the stability of the substrate at the active site needs to be studied. If the active site exists after a narrow tunnel, the basic understanding of the residues present in the tunnel becomes extremely important. To see the substrate binding-unbinding process, we need to implement the enhanced MD tools. Here, choosing of CV becomes extremely crucial. We can choose the distance between the centre of mass (COM) of the substrate and the COM of the binding residues. Multi CV is also implemented in recently advanced techniques like umbrella sampling, and Metadynamics using PLUMED. Along with distance, we can choose the basic dihedral, which is changing the whole substrate orientation. In every case, we must ensure that the chosen CVs should cover the slow dynamics of the binding-unbinding process. As in Quantumzyme, we mostly focus on enzyme engineering, to make enzyme work efficiently for the unnatural substrate. So enhanced sampling becomes critical technique to identify hotspots which tell us about interacting residue with ligand and there after mutate those residues along with screening the mutations based on free energy. CV is tremendously important to get reliable free energy landscape. So, with correct selection of CV/CVs we can identify the bottleneck of substrate binding to an enzyme.
Reference:
- Kästner, J. Umbrella Sampling. Wiley Interdiscip. Rev.: Comput. Mol. Sci. 2011, 1, 932-942.
- Torrie, Glenn M., and John P. Valleau. "Nonphysical sampling distributions in Monte Carlo free-energy estimation: Umbrella sampling." Journal of Computational Physics 23, no. 2 (1977): 187-199.
- Laio, Alessandro, and Francesco L. Gervasio. "Metadynamics: a method to simulate rare events and reconstruct the free energy in biophysics, chemistry and material science." Reports on Progress in Physics 71, no. 12 (2008): 126601
- Barducci, Alessandro, Massimiliano Bonomi, and Michele Parrinello. "Metadynamics." Wiley Interdisciplinary Reviews: Computational Molecular Science 1, no. 5 (2011): 826-843.