
Recombinant protein expression is the process of producing a specific protein in a host organism by introducing a recombinant gene into the host's cells. The gene of interest is expressed using recombinant DNA technology where the gene of interest is inserted into an expression vector and then transformed into the host cells. The transformed cells use the host cell machinery to transcribe and translate the protein of interest.
The host organisms used in the recombinant protein expression can be prokaryotic such as Escherichia coli (E. coli) or eukaryotic (yeast, insect cells, or mammalian cells) depending on the protein, the desired quantity, and the nature of post-translational modifications. E. coli as an expression system is the most preferred due to the following reasons. Firstly, it exhibits exceptionally rapid growth kinetics, with a doubling time of approximately 20 minutes under optimal conditions in glucose-salts media. Secondly, attaining high cell density cultures is easily achievable. Thirdly, rich complex media can be easily formulated from readily available and inexpensive components. Lastly, the transformation of E. coli with exogenous DNA is a fast and straightforward process. Plasmid transformation can be accomplished in as little as 5 minutes.
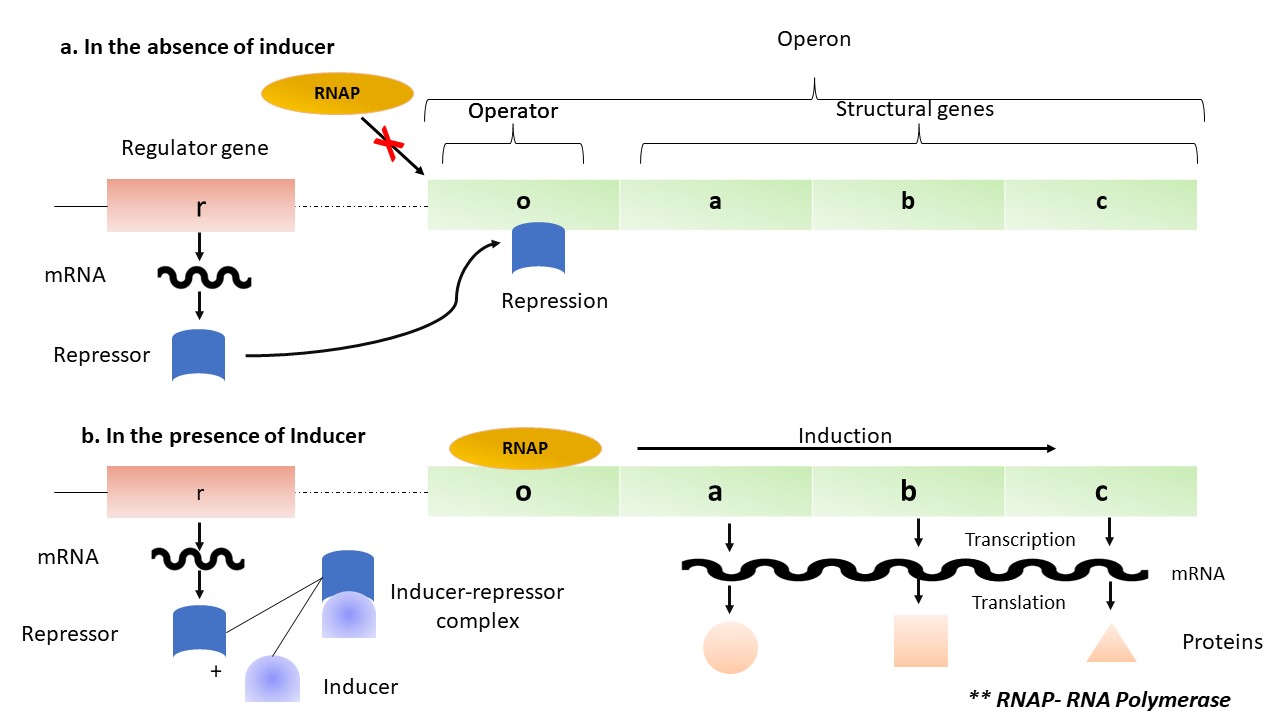
Fig 1. Schematic representation of operon
E. coli is a well-studied model organism for understanding prokaryotic genetics and molecular biology, including the concept of operons. An operon is a genetic unit consisting of a cluster of functionally related genes, typically involved in a specific metabolic pathway or biological function, which are transcribed together into a single mRNA molecule. In the absence of an inducer, repressor binds to the operator gene and inhibits RNA polymerase to bind to the operator gene and thereby inhibits the transcription and translation process. In the presence of inducer, an inducer-repressor complex is formed, which prevents the repressor from binding to the operator gene. This complex formation in turn allows the RNA polymerase to transcribe the gene of interest (Fig. 1).
- Induction strategies for overexpression of genes in E. coli
It is well established that inducers are required to initiate protein expression in the operons. This concept is being exploited by recombinant DNA technology for expression of the protein of interest. An optimization of parameters such as induction temperature, inducer concentration, incubation period after induction, etc are usually required to find the best conditions to reach the maximum expression level while avoiding cell damage. There are several strategies that can be used to induce the expression of genes in E. coli. Some of the most common strategies are discussed below:
- Induction by chemical molecules
Chemical inducers are small molecules that trigger the expression of specific genes by influencing the binding of regulatory proteins to the operator regions of the inducible operons. Some of the commonly used chemical inducers are lactose, IPTG and arabinose. The lac operon is a well-studied genetic regulatory system found in E. coli. It is a classic example of gene regulation and illustrates how organisms can efficiently control the expression of genes involved in specific metabolic pathways.
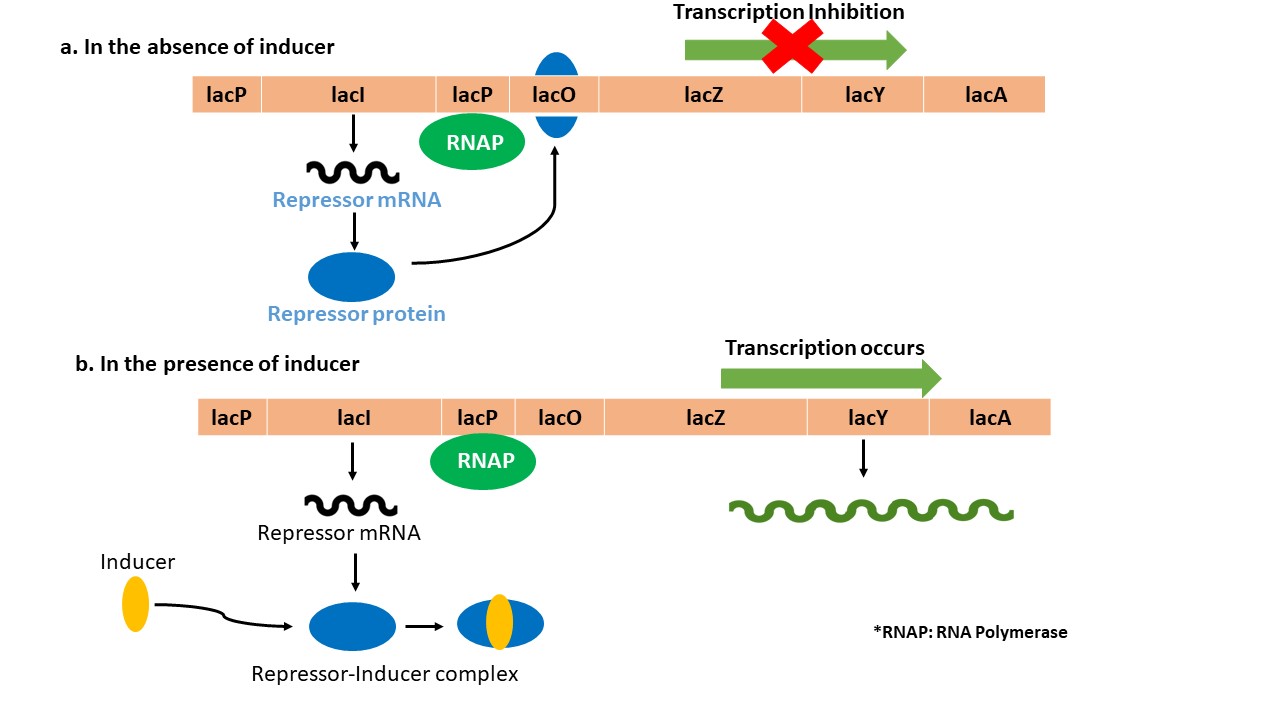
- Components of Lac operon
Structural Genes
lacZ, lacY, lacA are the three structural genes that encode for the enzyme beta-galactosidase, lactose permease and transacetylase respectively. These enzymes are responsible for the hydrolysis of lactose into glucose and galactose, isomerization of lactose into allolactose and facilitate the transport of lactose into the bacterial cell.
Promoter
The promoter (lacP) serves as the binding site for RNA polymerase, which initiates the transcription of the structural genes.
Operator
The operator (lacO) is located between the promoter and the structural genes. It controls the transcription of the structural genes by acting as the binding site for the lac repressor protein.
Repressor
The lac repressor (lacI) protein binds to the operator, preventing RNA polymerase from transcribing the structural genes in the absence of inducer.
- Mechanism of Lac operon
In the absence of IPTG/lactose, the repressor lacI binds to the operator lacO, preventing RNA polymerase from binding to the promoter lacP and inhibits transcription of structural genes – lacZ, lacY, lacA. In the presence of IPTG/lactose, the repressor binds to inducer molecule, thereby altering repressor conformation and prevents repressor binding to operator. Hence, RNA polymerase binds to the promoter and continues to transcribe the structural genes. IPTG is the most preferred inducer over lactose and allolactose as it is non-hydrolysable and hence, ensures continuous higher degree of control over the expression of target genes. Another advantage of IPTG is its high efficiency in small quantities to control gene expression. During recombinant expression of genes, it should be taken care that the recombinant plasmid must be transformed using a repressor negative (lacI-) E. coli strains eg: BL21(DE3), BL21(DE3)*, BL21(DE3) pLysS which allows effective the transcription of the target gene.
- Induction with temperature
Thermal induction is a very simple procedure with high productivity and avoids the use of expensive chemical inducers. However, a temperature change can occasionally activate the physiological responses such as heat shock proteins, chaperones and proteases, and SOS response. This system is particularly useful when studying genes that might be toxic to the host cell. One commonly used temperature-induced system in E. coli is the λ phage-based cI857 repressor system. It is based on the temperature sensitivity of a repressor protein derived from the bacteriophage lambda (λ).
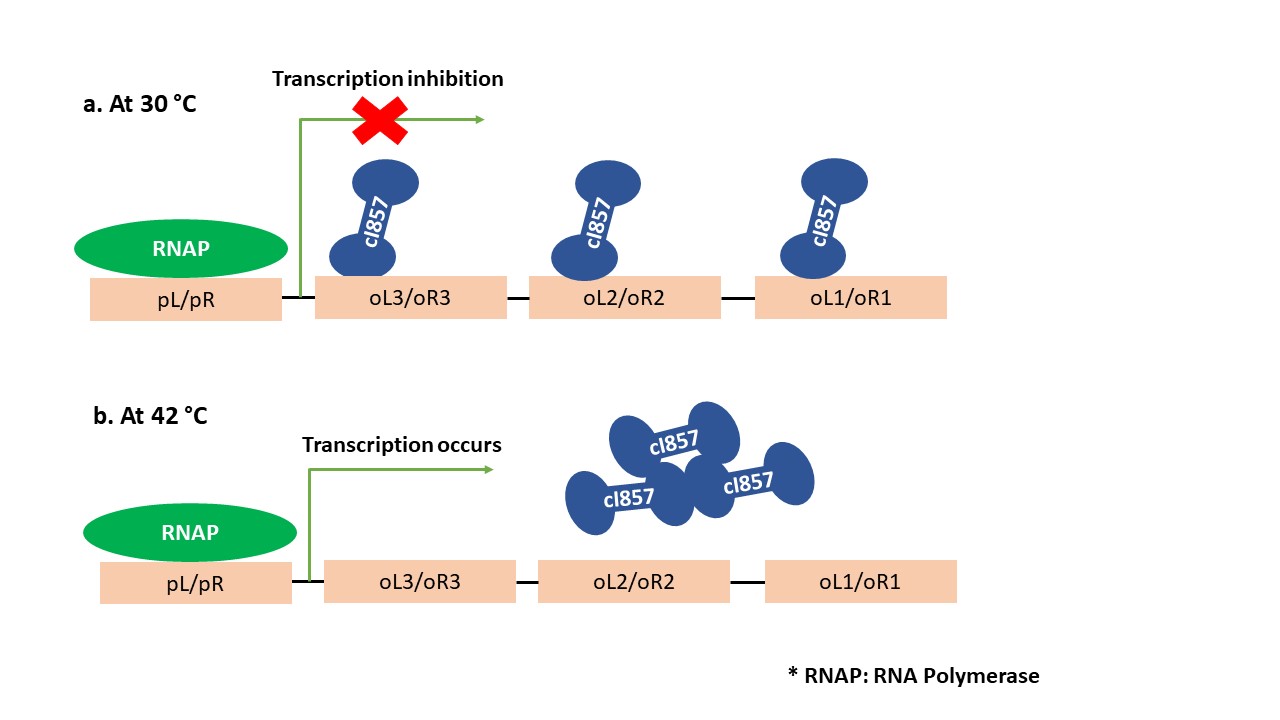
- Components of the cI857 operon system
- Repressor The cI857 repressor is a temperature-sensitive mutant of the λ phage repressor protein.
- Operator The cI857 repressor binds to the operator sites called oL/oR to control transcription.
- Promoter The cI857-regulated promoter - pL/pR is located upstream of the gene(s) to be controlled.
- Mechanism of cI857 Operon
At lower temperatures (25-30°C), the cI857 repressor forms dimers and engages with three operator regions (oL3/oR3, oL2/oR2, and oL1/oR1), leading to the prevention of RNA polymerase mediated transcription of the gene of interest positioned downstream of the pL or pR promoters. At higher temperatures (>40°C), the cI857 repressor's interaction with the oL or oR segments is prevented, allowing RNA polymerase-driven transcription to occur.
Figure 3. Schematic Representation temperature-controlled expression system.
- Induction with osmotic pressure
Osmotic stress is a physiological condition that occurs when there is an imbalance in the concentration of solutes (such as ions and molecules) between a cell and its surrounding environment. Inducing gene expression using sodium chloride (NaCl) in E. coli is a common approach, especially when studying genes involved in osmoregulation and stress response. The addition of NaCl to the growth medium can create conditions of high osmolarity, mimicking certain stress conditions. One well-known operon that responds to osmotic stress, is proU operon which is found in E. coli and related bacteria. It plays a crucial role in osmoregulation, particularly in response to high osmolarity caused by increased salt concentrations in the environment and helps to maintain cellular integrity.
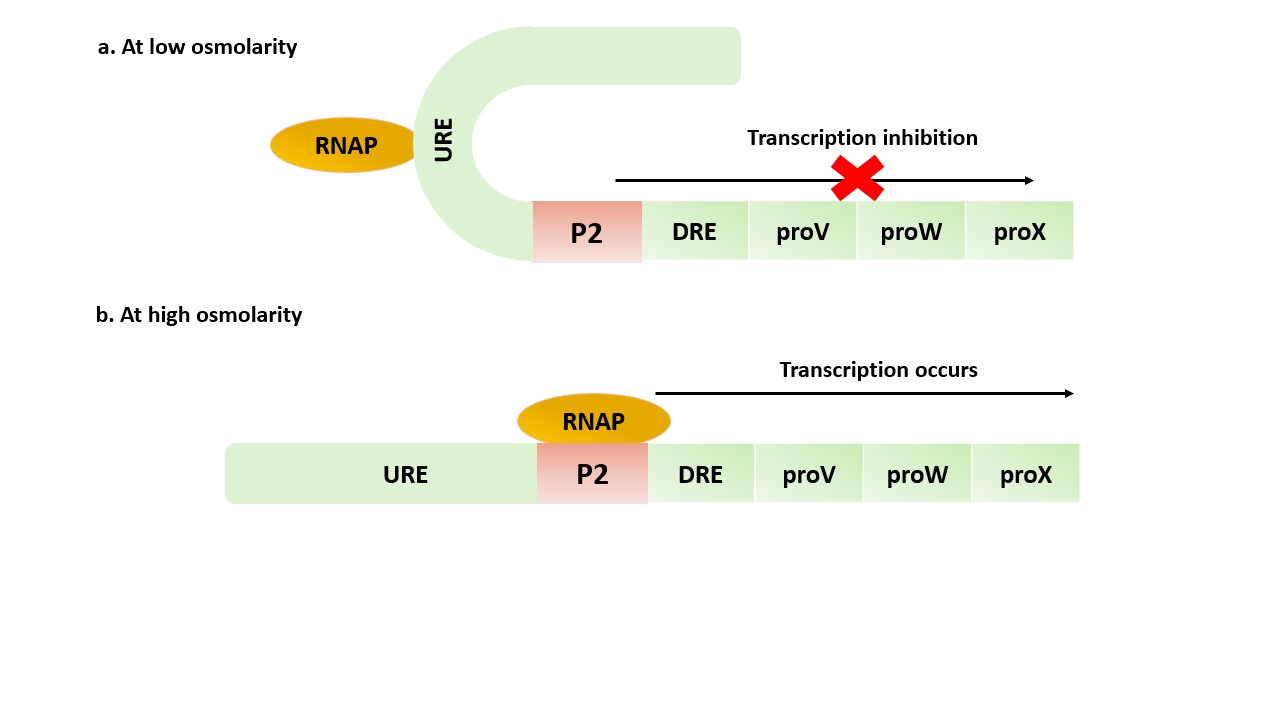
- Components of proU operon
- Structural genes
proV and proW genes encode for the ProV and ProW transmembrane proteins respectively. ProV forms a complex with ProW and is responsible for transporting compatible solutes across the cell membrane into the cell. proX gene encodes the ProX protein, which is a cytoplasmic protein and binds to the compatible solutes once they have been transported into the cell and hence, regulate solute accumulation and prevent toxicity to cell.
- Regulators
The regulatory region of the proU operon consists of the upstream regulatory element (URE) and a downstream regulatory element (DRE) positioned upstream and downstream of the proU P2 promoter respectively. Both URE and DRE contain high‐affinity histone-like nucleoid structuring (H-NS) protein binding sites.
- Mechanism of proU operon
At low osmolarity, increased interaction between URE-H-NS: DRE-H-NS happens, which prevents the binding of RNA polymerase to P2 and hence, prevent the transcription. At high osmolarity conditions - fewer contacts occur, thus weakening the URE: DRE interaction. This increases the accessibility of P2 for RNAP and thus increases the transcription efficiency.
Figure 4. Schematic representation of proU operon
- Enzyme production at Quantumzyme
At Quantumzyme™, we have successfully optimized and employed IPTG as an inducer for the expression of a wide range of enzymes. Currently, our focus is on refining the salt-based induction approach to eliminate the need for the expensive IPTG inducer, especially during large-scale enzyme production in fermentors. The salt-induced overexpression of the gene of interest is achieved using a specific E. coli strain known as GJ1158. This strain contains a single chromosomally integrated copy of the gene for phage T7 RNA polymerase, under the transcriptional control of the osmoresponsive proU operon. The concentration of salt (NaCl) added to the growth medium needs to be optimized for each protein, typically ranging from 0.1 M to 0.5 M.
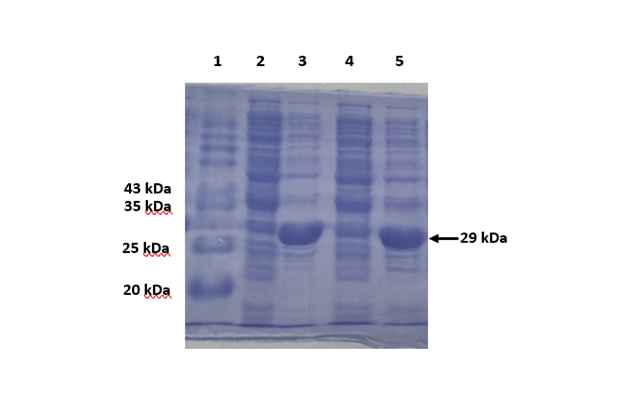
The SDS-PAGE gel below illustrates the NaCl-based induction method using the E. coli GJ1158 strain to express the QZ-ketoreductase gene, which is cloned in the pET28a vector. GJ1158, containing the ketoreductase gene, was cultured in LB media without NaCl. Growth was monitored until the OD600 reached 0.8, after which it was induced with 0.3 M NaCl. The expression was confirmed through SDS-PAGE (Figure 5). Enzyme expression with NaCl as the inducer was comparable to IPTG-based induction. NaCl-based induction offers several advantages, including being a cost-effective alternative inducer and reducing inclusion body formation.
5A. Expression Analysis
| Lane | Sample | Inducer |
| 1 | Protein Marker | NA |
| 2 | Uninduced | IPTG |
| 3 | Induced | IPTG |
| 4 | Uninduced | NaCl |
| 5 | Induced | NaCl |
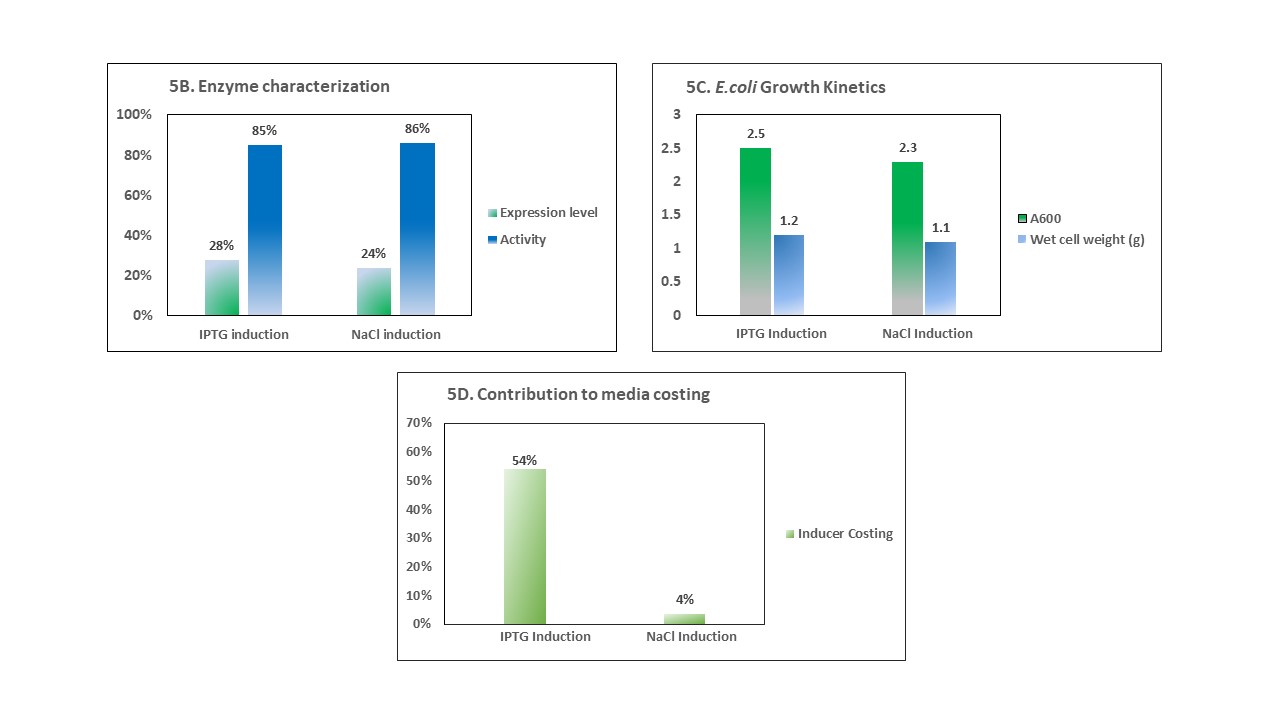
Figure 5. A. 12% SDS-PAGE depicting comparison of protein expression using IPTG and NaCl as inducer B-D. The graphs depict the comparison of enzyme parameters such as expression levels, enzyme activity, cell growth kinetics and the economic contribution of inducer between IPTG and NaCl based induction strategies. All the parameters are calculated to produce QZ-ketoreductase enzyme from 100 mL of recombinant E. coli culture.
As an enzyme engineering and biotransformation company, Quantumzyme™ is committed to constant innovation aimed at diminishing the economic footprint of industrial biocatalysis, advancing the principles of green chemistry, and consistently contributing to the scientific community through our ongoing research efforts.