Biocatalysis is the fastest growing technology for synthesis of chiral molecules. Quantumzyme’s biocatalysis offerings comprises of redox enzymes such as



| Description | Product Code | Accompanying documentation | Panel Contents* |
|---|---|---|---|
| Alcohol Dehydrogenase | ADH-001 | ADH Product guide Cofactor regeneration guide | 6 ADHs |
| Transaminase | TA-001 | TAM product guide Cofactor regeneration guide | 6 TAs |
| Diels-Alderase | DA-001 | DA product guide | 6 DAs |
The enzymes in this panel have been selected for their activity on a wideselection of substrates and in a widerange of pH, to encounter different synthetic needs of our customers.
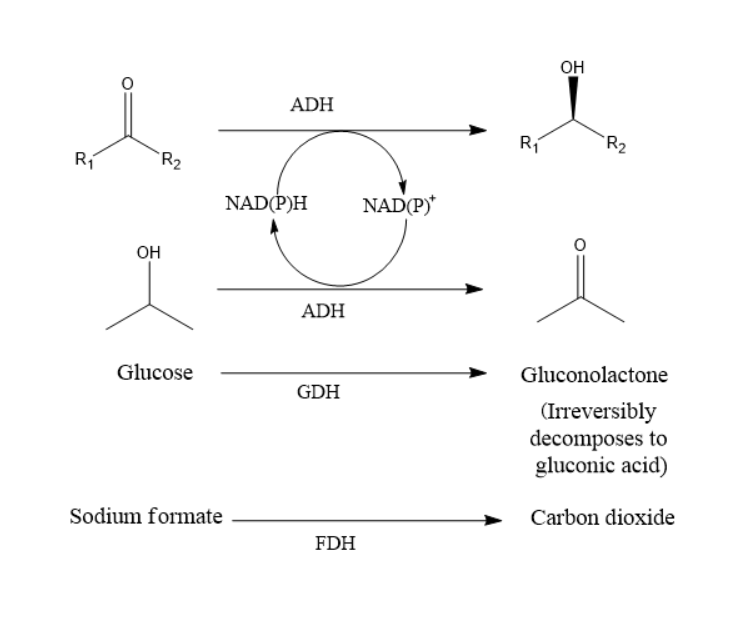
Enantiopure alcohols can be obtained using our ADHs. (R) and (S)-enantiomers of acetophenone and its derivatives can be accessed by using
these ADHs as well as racemic mixtures.
| Enzyme class | Enzyme | Cofactor regeneration | Optimum pH |
|---|---|---|---|
| Alcohol Dehydrogenase | ADH-001 | GDH, FDH | pH 7.5 - 8.0 |
| Alcohol Dehydrogenase> | ADH-002 | GDH, FDH | pH 7.5 - 8.0 |
| Alcohol Dehydrogenase | ADH-003 | GDH, FDH | pH 7.5 - 8.0 |
| Alcohol Dehydrogenase | ADH-004 | GDH, FDH | pH 7.5 - 8.0 |
| Alcohol Dehydrogenase | ADH-005 | GDH, FDH | pH 7.5 - 8.0 |
| Alcohol Dehydrogenase | ADH-006 | GDH, FDH | pH 7.5 - 8.0 |
The enzymes in this panel have been selected for their activity on a wideselection of substrates and in a widerange of pH, to encounter different synthetic needs of our customers.Aromatic and aliphatic primary amines can be obtained using our transaminases(TAs) enzymes for the formal reductive amination of ketone to the corresponding (R) or (S)-amine. The results will depend on the inherent enzyme substrate scope but also on the substrate properties.
In fact, if the amine moiety in the product molecule is stabilized by hydrogen bonds with other neighboring functionalities, the reaction will reach higher conversions because the concurrent deamination reaction is less favoured.

| Enzyme class | Enzyme | Cofactor regeneration | Optimum pH |
|---|---|---|---|
| Transaminase | TA -001 | GDH, FDH | pH 7.5 - 8.0 |
| Transaminase | TA-002 | GDH, FDH | pH 7.5 - 8.0 |
| Transaminase | TA-003 | GDH, FDH | pH 7.5 - 8.0 |
| Transaminase | TA-004 | GDH, FDH | pH 7.5 - 8.0 |
| Transaminase | TA-005 | GDH, FDH | pH 7.5 - 8.0 |
| Transaminase | TA-006 | GDH, FDH | pH 7.5 - 8.0 |
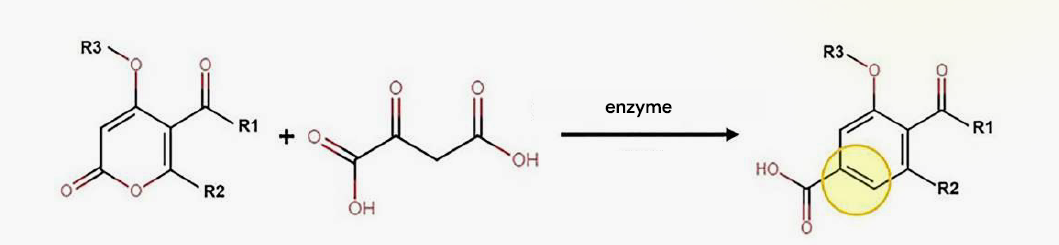
Diels-Alderase is an enzyme carrying out C-C bond formation between the diene and alkene/alkyne leading to cycloaddition reaction to form a cyclohexene ring. It requires MQ* ions for its activity. It works efficiently in the pH range of 7.0 -7.4 and temperature of 25°C 30°C.
| Shipping format | Enzyme purity | Enzyme concentration / amount | Pack size | Shipping condition | Storage |
|---|---|---|---|---|---|
| Liquid | Crude | 10 mg/mL | 5 | Dry ice | 15 |
| Liquid | Purified | 3 mg/mL | 3 | Dry ice | 20 |
| Freeze dried powder | Crude | 100 mg | 1 | Room Temperature | 20 |
| Freeze dried powder | Purified | 10 mg | 1 | Room Temperature | 25 |
Copyrights © 2023 All Rights Reserved by Quantumzyme LLP